Difference between revisions of "Module Vegetation"
From MohidWiki
Davidbrito (talk | contribs) (→If vegetation is readed from file) |
|||
| Line 106: | Line 106: | ||
=====leaf area index===== | =====leaf area index===== | ||
| + | Constant value, time serie, grid or HDF. To create one grid/hdf from various timeseries for different vegetation types see [[FillMatrix]]. | ||
| + | |||
| + | =====potential leaf area index===== | ||
Constant value, time serie, grid or HDF. To create one grid/hdf from various timeseries for different vegetation types see [[FillMatrix]]. | Constant value, time serie, grid or HDF. To create one grid/hdf from various timeseries for different vegetation types see [[FillMatrix]]. | ||
| Line 226: | Line 229: | ||
after senescence | after senescence | ||
NewLAI = LastLAIBeforeSenescence * LAIDecline - LAIGrazed - LAIRemovedInHarvest | NewLAI = LastLAIBeforeSenescence * LAIDecline - LAIGrazed - LAIRemovedInHarvest | ||
| + | |||
| + | =====potential leaf area index===== | ||
| + | This property is not simulated by the model so it has to be read. Constant value, time serie, grid or HDF. To create one grid/hdf from various timeseries for different vegetation types see [[FillMatrix]]. | ||
=====canopy height===== | =====canopy height===== | ||
Revision as of 17:14, 8 November 2010
Contents
Overview
Vegetation Model handles information about vegetation cover and interacts with atmosphere and soil properties. Vegetation dynamics can be handled by the model in two different manners: i) reading from file (time serie, hdf, grid); ii) using a vegetation growth model. The first option is the previous formulation where LAI and root depth properties are provided by user and water uptake is simulated. The second option uses a SWAT based vegetation growth model and plant biomass, LAI, nutrient content and nutrient uptake are explicitly simulated.
SWAT vegetation growth model uses the concepts from EPIC crop model (Izaurralde et al., 2006) of radiation-use efficiency by which a fraction of daily photosynthetically active radiation is intercepted by the plant canopy and converted into plant biomass. Gains in plant biomass are affected by vapor pressure deficits and atmospheric CO2 concentration. Stress indices for water, temperature, nitrogen, phosphorus and aeration are calculated using the value of the most severe of these stresses to reduce potential plant growth and crop yield. Nutrient uptake is done based on plant target (optimal content) and availability in soil.
Concepts
Property
Vegetation model was redesigned to be structured in properties instead of vegetation types. The advantage of this structure is that in the input file the number of properties is fixed (no matter the complexity of the vegetation cover) and input can be preprocessed for the entire grid (see How to pre-process vegetation). In the previous structure, applications with several vegetation covers could rapidly increase input file lines and input errors. More over as they are not graphed in time serie or hdf the visual inspection could take longer.
See the list of allowed properties names
Main processes
If Vegetation is not used
If the user chooses not to include vegetation in basin data file with the keyword:
VEGETATION : 0
then transpiration is not computed.
However, the user may want to still evaporate water from soil surface. To do so, evapotranspiration must be enabled in basin data file:
EVAPOTRANSPIRATION : 1
and reference evapotranspiration defined (property standard). In this case all the reference evapotranspiration will be in the form of potential evaporation.
If Vegetation is readed from file
If the user chooses to include vegetation in basin file with the keyword:
VEGETATION : 1
then transpiration is computed. If the user chooses the option to simulate vegetation giving properties evolution from file (hdf5, grid), than leaf area index, root depth, specific leaf storage and crop coefficient properties must be given. This option correspond to the old formulation and, yet, only water uptake is simulated.
Active Processes
If vegetation is read from file then water uptake and nutrient uptake may be modelled. Nutrient uptake can only be modelled if water uptake is.
WATER_STRESS : 0/1 !Connects/disconnects water uptake NITROGEN_STRESS : 0/1 !Connects/disconnects nitrogen uptake PHOSPHORUS_STRESS : 0/1 !Connects/disconnects phosphorus uptake
Water Uptake
This process corresponds to plant transpiration taking water from soil. Evapotranspiration must be enabled in basin data file:
EVAPOTRANSPIRATION : 1
The user may want to compute a global potential evapotranspiration or separate potential transpiration (in plants along the root depth) and potential evaporation (on soil surface) based on leaf area index. This option is defined in basin data file with the keyword:
EVAPOTRANSPIRATION_METHOD: 1/2 (1-Global Evapotranspiration; 2-Transpiration and Evaporation)
To use the read from file approach use the keyword in vegetation data file:
WATER_UPTAKE_METHOD : 1 (1- according to root profile; 2-SWAT based (exponential and tresholds)
Which means that the method for transpiration is the one from the formulation previous to the vegetation growth model.
Potential water uptake (potential evapotranspiration/transpiration) is distributed in depth (for each layer) according to root distribution with keyword in vegetation data file:
ROOT_PROFILE : 1/2 (1-Triangular; 2-Constant)
Water Uptake (actual uptake) computation takes in potential uptake and limits it to plant and soil constraints. Plant constraints can be done with two options: i) with Feddes formulation or ii) with van Genuchten curve with the keyword in vegetation data file:
WATER_UPTAKE_STRESS_METHOD: 1/2 (1-Feddes; 2-van Genuchten)
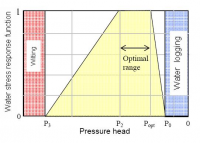
- Feddes formulation has plant tresholds. This means that plant has soil pressure heads tresholds where uptake is optimum and soil heads (under field capacity and below wilting point) where no uptake occurs. Between optimum and no transpiration linear interpolation is done:
Tp = Factor * PotentialTranspiration
where Tp is Effective transpiration in layer, Factor is the stress factor achieved for the layer and PotentialTranspiration is the PotentialTranspiration computed for the layer.
- van Genuchten formulation is an empirical curve also dependent on pressure head:
Insert Equation for van Genuchten
Water uptake is then limited to available water in soil (above residual content).
Additionally water uptake can be limited with soil velocity with the following keyword in vegetation data file:
LIMIT_TRANSP_WATER_VEL : 1
Soil pressure heads tresholds must be provided for each vegetation type in the vegetation file (example below):
!Arable Land - Trigo <beginvegetationtype> ID : 1 NAME : Agriculture FEDDES_H1 : -0.1 FEDDES_H2 : -0.25 FEDDES_H3 : -2.0 FEDDES_H4 : -80.0 <endvegetationtype> !Forest <beginvegetationtype> ID : 2 NAME : Forest FEDDES_H1 : -0.1 FEDDES_H2 : -0.25 FEDDES_H3 : -6.0 FEDDES_H4 : -30.0 <endvegetationtype>
Nutrient Uptake
Nutrient uptake may be done only using the formulation where the uptake mass is obtained from flow * concentration in layer.
Properties
Also Properties leaf area index, root depth, specific leaf storage and crop coefficent must be provided (from file or constant values). This must comply with fillmatrix standards under the <beginproperty> and <endpropery> blocks.
leaf area index
Constant value, time serie, grid or HDF. To create one grid/hdf from various timeseries for different vegetation types see FillMatrix.
potential leaf area index
Constant value, time serie, grid or HDF. To create one grid/hdf from various timeseries for different vegetation types see FillMatrix.
root depth
Constant value, time serie, grid or HDF. To create one grid/hdf from various timeseries for different vegetation types see FillMatrix.
specific leaf storage
Usually used constant value but applies the same as previous properties.
crop coefficient
Usually used constant value but applies the same as previous properties.
If Vegetation is explicitly simulated - Growth Model
If the user chooses to include vegetation in basin file with the keyword:
VEGETATION : 1
then transpiration is computed. If the user chooses the option to simulate explicitly vegetation, than plant biomass, root biomass, LAI, canopy height and nutrient content, are simulated.
Active Processes
If vegetation growth model is active then more options may be modelled.
WATER_STRESS : 0/1 !Connects/disconnects water limitation on plant growth NITROGEN_STRESS : 0/1 !Connects/disconnects nitrogen limitation on plant growth PHOSPHORUS_STRESS : 0/1 !Connects/disconnects phosphorus limitation on plant growth TEMPERATURE_STRESS : 0/1 !Connects/disconnects temperature limitation on plant growth ADJUST_RUE_FOR_CO2 : 0/1 !Connects/disconnects CO2 limitation on plant growth ADJUST_RUE_FOR_VPD : 0/1 !Connects/disconnects Vapour Pressure Deficit limitation on plant growth GRAZING : 0/1 !Connects/disconnects grazing MANAGEMENT : 0/1 !Connects/disconnects management DORMANCY : 0/1 !Connects/disconnects dormancy FERTILIZATION : 0/1 !Connects/disconnects fertilization
Water Uptake
This process corresponds to plant transpiration taking water from soil. Evapotranspiration must be enabled in basin data file:
EVAPOTRANSPIRATION : 1
The user may want to compute a global potential evapotranspiration or separate potential transpiration (in plants along the root depth) and potential evaporation (on soil surface) based on leaf area index. This option is defined in basin data file with the keyword:
EVAPOTRANSPIRATION_METHOD: 1/2 (1-Global Evapotranspiration; 2-Transpiration and Evaporation)
To use the vegetation growth model approach use the keyword in vegetation data file:
WATER_UPTAKE_METHOD : 2 (1- TP according to root profile; 2-SWAT based (TP exponential and tresholds))
Which means that the method for transpiration is the one from the vegetation growth model formulation.
Potential water uptake (potential evapotranspiration/transpiration) is distributed in depth according to a exponential distribution:
Insert potential water uptake distribution equation
Water Uptake (actual uptake) computation takes in potential uptake and limits it to soil constraints. consisting in water content in soil:
Insert low water content reduction
Insert high water content reduction (to do)
Nutrient Uptake
Nutrient uptake may be done in two ways, either using the SWAT formulation where it is disconnected from water uptake or using a new formulation that the uptake mass is obtained from flow * concentration in layer.
NUTRIENT_UPTAKE_METHOD : 1/2 !1- uptake is conc * water uptake; 2- SWAT based (independent of water uptake)
Also, nutrient stress may be computed either using SWAT formulation (relation to optimal and effective plant content) or using a new formulation where is the ratio between effective and optimal uptake (following water stress).
NUTRIENT_STRESS_METHOD : 1/2 !1- effective/optimal; 2- SWAT based
Optimal nutrient uptake is computed from plant optimal content and effective content
Equation for optimal nutrient content
Then, optimal nutrient uptake is distributed in depth similar to that of water
Equation for optimal uptake distribution with depth
Soil constraints are then taken into account because uptake is only allowed if enough mass exists in layer
Effective Uptake = min (Optimal Uptake, Available content in soil)
Properties
total plant biomass
Inside the property block choose:
EVOLUTION : 2 (property will be simulated with vegetation growth model)
Property evolution equation:
NewPlantBiomass = OldPlantBiomass + BiomassGrowth - BiomassGrazed - BiomassRemovedInHarvest
- BiomassRemovedInDormancy
total plant nitrogen
Inside the property block choose:
EVOLUTION : 2 (property will be simulated with vegetation growth model)
Property evolution equation:
NewPlantNitrogen = OldPlantNitrogen + NitrogenUptake - NitrogenGrazed - NitrogenRemovedInHarvest
- NitrogenRemovedInDormancy
total plant phosphorus
Inside the property block choose:
EVOLUTION : 2 (property will be simulated with vegetation growth model)
Property evolution equation:
NewPlantPhosphorus = OldPlantPhosphorus + PhosphorusUptake - PhosphorusGrazed
- PhosphorusRemovedInHarvest - PhosphorusRemovedInDormancy
root biomass
Inside the property block choose:
EVOLUTION : 2 (property will be simulated with vegetation growth model)
Property evolution equations:
RootBiomass = RootFraction * PlantBiomass
RootFraction = 0.4 - 0.2 * HUAccumulated
root depth
Inside the property block choose:
EVOLUTION : 2 (property will be simulated with vegetation growth model)
Property evolution equations: for annuals, legumes
RootDepth = 2.5 * HUAccumulated * MaxRootDepth
for trees, perennials
RootDepth = MaxRootDepth
leaf area index
Inside the property block choose:
EVOLUTION : 2 (property will be simulated with vegetation growth model)
Property evolution equations: before senescence
NewLAI = OldLAI + LAIGrowth - LAIGrazed - LAIRemovedInHarvest
after senescence
NewLAI = LastLAIBeforeSenescence * LAIDecline - LAIGrazed - LAIRemovedInHarvest
potential leaf area index
This property is not simulated by the model so it has to be read. Constant value, time serie, grid or HDF. To create one grid/hdf from various timeseries for different vegetation types see FillMatrix.
canopy height
Inside the property block choose:
EVOLUTION : 2 (property will be simulated with vegetation growth model)
Property evolution equation:
CanopyHeight = MaxCanopyHeight * SQRT(MaxLAIfraction)
specific leaf storage
This property is not simulated by the model so it has to be read. Usually is a constant value but it can be defined also as timeserie, grid or HDF. To create one grid/hdf from various timeseries for different vegetation types see FillMatrix.
Inside the property block choose:
EVOLUTION : 1 (property will be read by the model)
crop coefficient
This property is not simulated by the model so it has to be read. Usually is a constant value but it can be defined also as timeserie, grid or HDF. To create one grid/hdf from various timeseries for different vegetation types see FillMatrix.
Inside the property block choose:
EVOLUTION : 1 (property will be read by the model)
Other Features
How to generate the vegetation grid
A vegetation grid must be provided. One possible option is to extract info from land use shape file. In this case can use MOHID GIS going to menu [Tools]->[Shape to Grid Data] and provide: i) the grid (model grid), ii) the land use shape file and iii) the corespondence between land use codes and vegetation ID. In vegetation data file define the just created grid:
VEGETATION_ID_FILE : ..\..\GeneralData\Vegetation\VegetationID.dat
Define vegetation properties
Vegetation properties may be read or simulated according to the below.
This vegetation properties have to be given (not simulated):
- specific leaf storage
- crop coefficient
This vegetation properties may be given or simulated:
- leaf area index
- root depth
This vegetation properties may be simulated:
- total plant biomass
- total plant nitrogen
- total plant phosphorus
- root biomass
- canopy height
Properties are defined accordingly with Module FillMatrix standards in the block:
<beginproperty> NAME : root depth UNITS : m DESCRIPTION : plant root depth EVOLUTION : 1 OLD : 0 FILE_IN_TIME : NONE INITIALIZATION_METHOD : CONSTANT DEFAULTVALUE : 0.2 REMAIN_CONSTANT : 1 OUTPUT_HDF : 0 TIME_SERIE : 1 BOX_TIME_SERIE : 0 <endproperty>
EVOLUTION keyword value : 1 means that is not simulated but read. In other end EVOLUTION keyword value : 2 means that the property is simulated.
Simulation Options
If you decide not to use vegetation growth model (and instead, the old formulation) than the only needed properties (values must be provided) are:
- specific leaf storage
- crop coefficient
- leaf area index
- root depth
In the other end, if you decide to use vegetation growh model these properties may be used (model simulates them)
- leaf area index
- root depth
- total plant biomass
- total plant nitrogen (if simulating nitrogen)
- total plant phosphorus (if simulating phosphorus)
- root biomass
- canopy height
and only:
- specific leaf storage
- crop coefficient
need to have values defined for the simulation. All other properties will be simulated.
How to pre-process vegetation properties
Fill Matrix was updated to fill grids without interpolation. Instead of space stations (X,Y coordinate) user has to provide a vegetation grid with ID's and the values assigned to each ID (time serie or single value). Fill Matrix reads ID in the grid and searches for the value to fill the cell. If cells are not filled error message is sent. See FillMatrix for more details
Outputs
Time series
To write time series results define keyword:
TIME_SERIE : 1
in each property that you wish to write results.
Box integration
Maps (HDF5 format)
To write 3D results use keyword OUTPUT_TIME and define keyword:
OUTPUT_HDF : 1
in each property that you wish to write results.
Statistics
References
Izaurralde, R.C.; Williams, J.R. ; McGill, W.B.; Rosenberg, N.J.; Quiroga Jakas, M.C. (2006) - Simulating soil C dynamics with EPIC: Model description and testing against long‐term data. Ecol. Model. 192(3‐4): 362‐384.
Data File
Keywords
[Keyword] [Format] [Units] [Default] [Short Description]
VEGETATION_ID_FILE : string - [-] !Vegetation distribution grid path
VEGETATION_DT : real s [86400.] !Vegetation DT
INTEGRATION_DT : real s [ModelDT] !DT to integrate external variables until vegetation is
! is called (vegetation DT)
WATER_STRESS : 0/1 - [1] !Connects/disconnects water limitation on plant growth?
NITROGEN_STRESS : 0/1 - [1] !Connects/disconnects nitrogen limitation on plant growth?
PHOSPHORUS_STRESS : 0/1 - [1] !Connects/disconnects phosphorus limitation on plant growth?
TEMPERATURE_STRESS : 0/1 - [1] !Connects/disconnects temperature limitation on plant growth?
ADJUST_RUE_FOR_CO2 : 0/1 - [1] !Connects/disconnects CO2 limitation on plant growth?
ADJUST_RUE_FOR_VPD : 0/1 - [1] !Connects/disconnects Vapour Pressure Deficit limitation on
plant growth?
GRAZING : 0/1 - [0] !Connects/disconnects grazing
MANAGEMENT : 0/1 - [0] !Connects/disconnects management
DORMANCY : 0/1 - [0] !Connects/disconnects dormancy
FERTILIZATION : 0/1 - [0] !Connects/disconnects fertilization
NUTRIENT_FLUXES_WITH_SOIL : 0/1 - [1] !Connects/disconnects nutrient fluxes with soil
WATER_UPTAKE_METHOD : integer - [1] !1- according to root profile; 2-SWAT based (exponential
and tresholds)
LIMIT_TRANSP_WATER_VEL : 0/1 - [0] !Read if TRANSPIRATION_METHOD == 1.
ROOT_PROFILE : integer - [1] !Read if TRANSPIRATION_METHOD == 1:
!1-Triangular; 2-Constant; 3-Exponential(SWAT like)
WATER_UPTAKE_STRESS_METHOD : integer - [1] !Read if TRANSPIRATION_METHOD == 1: 1-Feddes; 2-VanGenuchten
NUTRIENT_UPTAKE_METHOD : integer - [2] !1- uptake is: conc * water uptake; 2- SWAT based
(independent of water uptake)
NUTRIENT_STRESS_METHOD : integer - [2] !1- effective/optimal; 2- SWAT based
CHANGE_LAI_SENESCENCE : 0/1 - [0] !Changes made to swat code because showed error with
CHANGE_CANOPY_HEIGHT : 0/1 - [0] grazing
ATMOSPHERE_OUTPUT : 0/1 - [0] !Output averaged atmosphere properties during dt
FLUXES_TO_SOIL_OUTPUT : 0/1 - [0] !Output fluxes to soil
ATMOSPHERE_CO2 : real ppm [330.] !Atmosphere CO2 concetrations - should be atmosphere property
WATER_UPTAKE_COMPENSATION_FACTOR : real - [0.] !Factor for uptake compensation from lower layers if computed
!layer demand is not met
!If zero there will exist no compensation. If 1. total demand
!no met may come from lower layers
NITROGEN_DISTRIBUTION_PARAMETER : real [-20.]
PHOSPHORUS_DISTRIBUTION_PARAMETER: real [-20.]
<beginproperty>
See module fillmatrix
EVOLUTION : integer - 1 !Property evolution: 1-Read from file
!2-vegetation growth model
<endproperty>
<beginvegetationtype>
ID
NAME
HAS_LEAVES
FEDDES_H1
FEDDES_H2
FEDDES_H3
FEDDES_H4
<begintimingdatabase>
MATURITY_HU : 1700.
PLANTING_JULIANDAY : -99.
PLANTING_HUBASE : 0.15
<endgtimingdatabase>
<begingrowthdatabase>
PLANT_TYPE : 5
OPTIMAL_NITROGENFRACTION_N1 : 0.0663
OPTIMAL_NITROGENFRACTION_N2 : 0.0255
OPTIMAL_NITROGENFRACTION_N3 : 0.0148
OPTIMAL_PHOSPHORUSFRACTION_P1 : 0.0053
OPTIMAL_PHOSPHORUSFRACTION_P2 : 0.0020
OPTIMAL_PHOSPHORUSFRACTION_P3 : 0.0012
BASE_TEMPERATURE : 0.
OPTIMAL_TEMPERATURE : 18.0
RADIATION_EXTINCTION_COEF : 0.65
BIOMASS_ENERGY_RATIO : 30.0
CO2_HIGH : 660.0
BIOMASS_ENERGY_RATIO_HIGH : 39.0
RUE_DECLINE_RATE : 6.0
LAI_MAX : 4.0
OPTIMAL_LAIMAXFRACTION_1 : 0.05
OPTIMAL_LAIMAXFRACTION_2 : 0.95
GROWFRACTION_1 : 0.05
GROWFRACTION_2 : 0.45
GROWFRACTION_LAIDECLINE : 0.50
ROOT_DEPTH_MAX : 1.30
CANOPY_HEIGHT_MAX : 0.9
OPTIMAL_HARVEST_INDEX : 0.4
MINIMUM_HARVEST_INDEX : 0.2
YELD_NITROGENFRACTION : 0.0250
YELD_PHOSPHORUSFRACTION : 0.0022
<endgrowthdatabase>
<beginmanagementandgrazedatabase>
GRAZING_START_JULIANDAY : -99.
GRAZING_START_PLANTHU : 0.5
GRAZING_DAYS : 10
MINIMUM_BIOMASS_FOR_GRAZING : 10.
GRAZING_BIOMASS : 70.
TRAMPLING_BIOMASS : 30.
HARVESTKILL_JULIANDAY : -99.
HARVESTKILL_PLANTHU : 1.2
HARVEST_JULIANDAY : -99.
HARVEST_PLANTHU : -99.
HARVEST_EFFICIENCY : 1.0
KILL_JULIANDAY : -99.
KILL_PLANTHU : -99.
<endmanagementandgrazedatabase>
<beginfertilizationdatabase>
MINERAL_N_FRACTION_IN_FERTILIZER : -99.
ORGANIC_N_FRACTION_IN_FERTILIZER : -99.
AMMONIA_FRACTION_IN_MINERAL_N : -99.
MINERAL_P_FRACTION_IN_FERTILIZER : -99.
ORGANIC_P_FRACTION_IN_FERTILIZER : -99.
FERTILIZER_FRACTION_IN_SURFACE : -99.
!!beginautofertilization!!
NITROGEN_TRESHOLD : -99.
NITROGEN_APPLICATION_MAX : -99.
NITROGEN_ANNUAL_MAX : -99.
EXPLICIT_PHOSPHORUS : 0
PHOSPHORUS_TRESHOLD : -99.
PHOSPHORUS_APPLICATION_MAX : -99.
PHOSPHORUS_ANNUAL_MAX : -99.
!!endautofertilization!!
!!beginscheduledfertilization!!
FERTILIZATION_JULIANDAY :-99.
FERTILIZATION_HU :-99.
!!endscheduledfertilization!!
<endfertilizationdatabase>
<endvegetationtype>
Sample
If vegetation is readed from file
WATER_STRESS : 1 NITROGEN_STRESS : 0 PHOSPHORUS_STRESS : 0 WATER_UPTAKE_METHOD : 1 !1- TP according to root profile ROOT_PROFILE : 1 !1- triangular; 2- Constant; 3-Exponential (only read if WATER_UPTAKE_METHOD : 1) WATER_UPTAKE_STRESS_METHOD : 1 !1-Feddes; 2- VanGenuchten (only read if WATER_UPTAKE_METHOD : 1) TIME_SERIE_LOCATION : ..\..\GeneralData\TimeSeriesLocation.dat VEGETATION_ID_FILE : ..\..\GeneralData\vegetation.dat OUTPUT_TIME : 0. 86400. <beginproperty> NAME : root depth UNITS : m DESCRIPTION : plant root depth EVOLUTION : 1 OLD : 0 FILE_IN_TIME : HDF FILENAME : ..\..\GeneralData\Trancao\Vegetation\RootDepth.hdf5 DEFAULTVALUE : 0.0 REMAIN_CONSTANT : 0 OUTPUT_HDF : 1 TIME_SERIE : 1 BOX_TIME_SERIE : 0 <endproperty> <beginproperty> NAME : leaf area index UNITS : m2/m2 DESCRIPTION : plant leaf area index EVOLUTION : 1 OLD : 0 FILE_IN_TIME : HDF FILENAME : ..\..\GeneralData\Trancao\Vegetation\LAI.hdf5 DEFAULTVALUE : 0. REMAIN_CONSTANT : 0 OUTPUT_HDF : 1 TIME_SERIE : 1 BOX_TIME_SERIE : 0 <endproperty> <beginproperty> NAME : specific leaf storage UNITS : m3/m2 DESCRIPTION : plant specific leaf storage EVOLUTION : 1 OLD : 0 FILE_IN_TIME : HDF FILENAME : ..\..\GeneralData\Trancao\Vegetation\SpecificLeafStorage.hdf5 DEFAULTVALUE : 0.0001 REMAIN_CONSTANT : 0 OUTPUT_HDF : 0 TIME_SERIE : 1 BOX_TIME_SERIE : 0 <endproperty> <beginproperty> NAME : crop coefficient UNITS : - DESCRIPTION : plant transpiration coefficient EVOLUTION : 1 OLD : 0 FILE_IN_TIME : HDF FILENAME : ..\..\GeneralData\Trancao\Vegetation\CropCoefficient.hdf5 DEFAULTVALUE : 1. REMAIN_CONSTANT : 0 OUTPUT_HDF : 0 TIME_SERIE : 1 BOX_TIME_SERIE : 0 <endproperty> !Arable Land - Trigo <beginvegetationtype> ID : 1 NAME : Agriculture FEDDES_H1 : -0.1 FEDDES_H2 : -0.25 FEDDES_H3 : -2.0 FEDDES_H4 : -80.0 <endvegetationtype> !Floresta <beginvegetationtype> ID : 2 NAME : Forest FEDDES_H1 : -0.1 FEDDES_H2 : -0.25 FEDDES_H3 : -6.0 FEDDES_H4 : -30.0 <endvegetationtype>
If vegetation growth model is used
VEGETATION_ID_FILE : ..\..\GeneralData\vegetation_2cells.dat VEGETATION_DT : 86400. INTEGRATION_DT : 3600. !DT to integrate atmosphere properties WATER_STRESS : 1 NITROGEN_STRESS : 0 PHOSPHORUS_STRESS : 0 TEMPERATURE_STRESS : 0 ADJUST_RUE_FOR_CO2 : 0 ADJUST_RUE_FOR_VPD : 0 GRAZING : 0 MANAGEMENT : 1 DORMANCY : 1 FERTILIZATION : 0 NUTRIENT_FLUXES_WITH_SOIL : 0 WATER_UPTAKE_METHOD : 2 !1- TP according to root profile; 2-SWAT based (TP exponential and tresholds) LIMIT_TRANSP_WATER_VEL : 0 ROOT_PROFILE : 1 !1- triangular; 2- Constant; 3-Exponential (only read if WATER_UPTAKE_METHOD : 1) WATER_UPTAKE_STRESS_METHOD : 1 !1-Feddes; 2- VanGenuchten (only read if WATER_UPTAKE_METHOD : 1) NUTRIENT_UPTAKE_METHOD : 2 !1- uptake is conc * water uptake; 2- SWAT based (independent of water uptake) NUTRIENT_STRESS_METHOD : 1 !1- effective/optimal; 2- SWAT based TIME_SERIE_LOCATION : ..\..\GeneralData\TimeSeriesLocation2D_2.dat OUTPUT_TIME : 0. 86400. ATMOSPHERE_OUTPUT : 1 FLUXES_TO_SOIL_OUTPUT : 1 !Potential total HU (yearly HU) - SUMi=1to12(average monthly temperature in month i * days in month i) <begin_TotalPotentialHU> INITIALIZATION_METHOD : CONSTANT DEFAULTVALUE : 5475. REMAIN_CONSTANT : 1 <end_TotalPotentialHU> <beginproperty> NAME : total plant biomass UNITS : kg/ha DESCRIPTION : plant biomass EVOLUTION : 2 OLD : 0 !DEFAULTVALUE : 1000. DEFAULTVALUE : 0. REMAIN_CONSTANT : 0 OUTPUT_HDF : 1 TIME_SERIE : 1 BOX_TIME_SERIE : 0 <endproperty> !<beginproperty> NAME : total plant nitrogen UNITS : kg/ha DESCRIPTION : plant nitrogen content EVOLUTION : 2 OLD : 0 !DEFAULTVALUE : 50. DEFAULTVALUE : 0. REMAIN_CONSTANT : 0 OUTPUT_HDF : 1 TIME_SERIE : 1 BOX_TIME_SERIE : 0 !<endproperty> !<beginproperty> NAME : total plant phosphorus UNITS : kg/ha DESCRIPTION : plant phosphorus content EVOLUTION : 2 OLD : 0 FILE_IN_TIME : NONE INITIALIZATION_METHOD : CONSTANT !DEFAULTVALUE : 1. DEFAULTVALUE : 0. REMAIN_CONSTANT : 0 OUTPUT_HDF : 1 TIME_SERIE : 1 BOX_TIME_SERIE : 0 !<endproperty> <beginproperty> NAME : root biomass UNITS : kg/ha DESCRIPTION : plant root biomass EVOLUTION : 2 OLD : 0 FILE_IN_TIME : NONE INITIALIZATION_METHOD : CONSTANT !DEFAULTVALUE : 200. DEFAULTVALUE : 0. REMAIN_CONSTANT : 0 OUTPUT_HDF : 1 TIME_SERIE : 1 BOX_TIME_SERIE : 0 <endproperty> <beginproperty> NAME : root depth UNITS : m DESCRIPTION : plant root depth EVOLUTION : 2 OLD : 0 FILE_IN_TIME : NONE INITIALIZATION_METHOD : CONSTANT !DEFAULTVALUE : 1. DEFAULTVALUE : 0.0 REMAIN_CONSTANT : 0 OUTPUT_HDF : 1 TIME_SERIE : 1 BOX_TIME_SERIE : 0 <endproperty> <beginproperty> NAME : leaf area index UNITS : m2/m2 DESCRIPTION : plant leaf area index EVOLUTION : 2 OLD : 0 FILE_IN_TIME : NONE !FILENAME : ..\..\GeneralData\LAI-2001-2007-RZWQM.dat !DATA_COLUMN : 2 DEFAULTVALUE : 0. REMAIN_CONSTANT : 0 OUTPUT_HDF : 1 TIME_SERIE : 1 BOX_TIME_SERIE : 0 <endproperty> <beginproperty> NAME : canopy height UNITS : m DESCRIPTION : plant canopy height EVOLUTION : 2 OLD : 0 FILE_IN_TIME : NONE INITIALIZATION_METHOD : CONSTANT DEFAULTVALUE : 0. REMAIN_CONSTANT : 0 OUTPUT_HDF : 1 TIME_SERIE : 1 BOX_TIME_SERIE : 0 <endproperty> <beginproperty> NAME : specific leaf storage UNITS : m3/m2 DESCRIPTION : plant specific leaf storage EVOLUTION : 1 OLD : 0 FILE_IN_TIME : NONE INITIALIZATION_METHOD : CONSTANT DEFAULTVALUE : 0.0001 REMAIN_CONSTANT : 1 OUTPUT_HDF : 0 TIME_SERIE : 1 BOX_TIME_SERIE : 0 <endproperty> <beginproperty> NAME : crop coefficient UNITS : - DESCRIPTION : plant transpiration coefficient EVOLUTION : 1 OLD : 0 FILE_IN_TIME : NONE INITIALIZATION_METHOD : CONSTANT DEFAULTVALUE : 1. REMAIN_CONSTANT : 1 OUTPUT_HDF : 0 TIME_SERIE : 1 BOX_TIME_SERIE : 0 <endproperty> !Arable Land - Trigo <beginvegetationtype> ID : 1 NAME : Agriculture HAS_LEAVES : 1 <begintimingdatabase> PLANTING_JULIANDAY : -99. PLANTING_HUBASE : 0.15 MATURITY_HU : 1700. <endtimingdatabase> <begingrowthdatabase> PLANT_TYPE : 5 OPTIMAL_NITROGENFRACTION_N1 : 0.0663 OPTIMAL_NITROGENFRACTION_N2 : 0.0255 OPTIMAL_NITROGENFRACTION_N3 : 0.0148 OPTIMAL_PHOSPHORUSFRACTION_P1 : 0.0053 OPTIMAL_PHOSPHORUSFRACTION_P2 : 0.0020 OPTIMAL_PHOSPHORUSFRACTION_P3 : 0.0012 BASE_TEMPERATURE : 0. OPTIMAL_TEMPERATURE : 18.0 RADIATION_EXTINCTION_COEF : 0.65 BIOMASS_ENERGY_RATIO : 30.0 CO2_HIGH : 660.0 BIOMASS_ENERGY_RATIO_HIGH : 39.0 RUE_DECLINE_RATE : 6.0 LAI_MAX : 4.0 OPTIMAL_LAIMAXFRACTION_1 : 0.05 OPTIMAL_LAIMAXFRACTION_2 : 0.95 GROWFRACTION_1 : 0.05 GROWFRACTION_2 : 0.45 GROWFRACTION_LAIDECLINE : 0.50 ROOT_DEPTH_MAX : 1.30 CANOPY_HEIGHT_MAX : 0.9 OPTIMAL_HARVEST_INDEX : 0.4 MINIMUM_HARVEST_INDEX : 0.2 YELD_NITROGENFRACTION : 0.0250 YELD_PHOSPHORUSFRACTION : 0.0022 <endgrowthdatabase> <beginmanagementandgrazedatabase> GRAZING_START_JULIANDAY : -99. GRAZING_START_PLANTHU : 0.5 GRAZING_DAYS : 10 MINIMUM_BIOMASS_FOR_GRAZING : 10. GRAZING_BIOMASS : 70. TRAMPLING_BIOMASS : 30. HARVESTKILL_JULIANDAY : -99. HARVESTKILL_PLANTHU : 1.2 HARVEST_JULIANDAY : -99. HARVEST_PLANTHU : -99. HARVEST_EFFICIENCY : 1.0 KILL_JULIANDAY : -99. KILL_PLANTHU : -99. <endmanagementandgrazedatabase> <beginfertilizationdatabase> MINERAL_N_FRACTION_IN_FERTILIZER : -99. ORGANIC_N_FRACTION_IN_FERTILIZER : -99. AMMONIA_FRACTION_IN_MINERAL_N : -99. MINERAL_P_FRACTION_IN_FERTILIZER : -99. ORGANIC_P_FRACTION_IN_FERTILIZER : -99. FERTILIZER_FRACTION_IN_SURFACE : -99. !!beginautofertilization!! NITROGEN_TRESHOLD : -99. NITROGEN_APPLICATION_MAX : -99. NITROGEN_ANNUAL_MAX : -99. EXPLICIT_PHOSPHORUS : 0 PHOSPHORUS_TRESHOLD : -99. PHOSPHORUS_APPLICATION_MAX : -99. PHOSPHORUS_ANNUAL_MAX : -99. !!endautofertilization!! !!beginscheduledfertilization!! FERTILIZATION_JULIANDAY :-99. FERTILIZATION_HU :-99. !!endscheduledfertilization!! <endfertilizationdatabase> <endvegetationtype> !Floresta <beginvegetationtype> ID : 2 NAME : Forest HAS_LEAVES : 1 <begintimingdatabase> PLANTING_JULIANDAY : -99. PLANTING_HUBASE : 0.15 MATURITY_HU : 1700. <endtimingdatabase> <begingrowthdatabase> PLANT_TYPE : 7 OPTIMAL_NITROGENFRACTION_N1 : 0.0060 OPTIMAL_NITROGENFRACTION_N2 : 0.0020 OPTIMAL_NITROGENFRACTION_N3 : 0.0015 OPTIMAL_PHOSPHORUSFRACTION_P1 : 0.0007 OPTIMAL_PHOSPHORUSFRACTION_P2 : 0.0004 OPTIMAL_PHOSPHORUSFRACTION_P3 : 0.0003 BASE_TEMPERATURE : 10. OPTIMAL_TEMPERATURE : 30.0 RADIATION_EXTINCTION_COEF : 0.65 BIOMASS_ENERGY_RATIO : 15.0 CO2_HIGH : 660.0 BIOMASS_ENERGY_RATIO_HIGH : 16.0 RUE_DECLINE_RATE : 8.0 LAI_MAX : 5.0 OPTIMAL_LAIMAXFRACTION_1 : 0.05 OPTIMAL_LAIMAXFRACTION_2 : 0.95 GROWFRACTION_1 : 0.05 GROWFRACTION_2 : 0.40 GROWFRACTION_LAIDECLINE : 0.99 ROOT_DEPTH_MAX : 3.50 CANOPY_HEIGHT_MAX : 6.0 OPTIMAL_HARVEST_INDEX : 0.76 MINIMUM_HARVEST_INDEX : 0.010 YELD_NITROGENFRACTION : 0.0015 YELD_PHOSPHORUSFRACTION : 0.0003 TREE_YEARSTOMATURITY : -99. TREE_MAXIMUMBIOMASS : -99. <endgrowthdatabase> <beginmanagementandgrazedatabase> GRAZING_START_JULIANDAY : -99. GRAZING_START_PLANTHU : -99. GRAZING_DAYS : 0. MINIMUM_BIOMASS_FOR_GRAZING : 0. GRAZING_BIOMASS : 0. TRAMPLING_BIOMASS : 0. HARVESTKILL_JULIANDAY : -99. HARVESTKILL_PLANTHU : -99. HARVEST_JULIANDAY : -99. HARVEST_PLANTHU : -99. HARVEST_EFFICIENCY : 1.0 KILL_JULIANDAY : -99. KILL_PLANTHU : -99. <endmanagementandgrazedatabase> <beginfertilizationdatabase> MINERAL_N_FRACTION_IN_FERTILIZER : -99. ORGANIC_N_FRACTION_IN_FERTILIZER : -99. AMMONIA_FRACTION_IN_MINERAL_N : -99. MINERAL_P_FRACTION_IN_FERTILIZER : -99. ORGANIC_P_FRACTION_IN_FERTILIZER : -99. FERTILIZER_FRACTION_IN_SURFACE : -99. !!beginautofertilization!! NITROGEN_TRESHOLD : -99. NITROGEN_APPLICATION_MAX : -99. NITROGEN_ANNUAL_MAX : -99. EXPLICIT_PHOSPHORUS : 0 PHOSPHORUS_TRESHOLD : -99. PHOSPHORUS_APPLICATION_MAX : -99. PHOSPHORUS_ANNUAL_MAX : -99. !!endautofertilization!! !!beginscheduledfertilization!! FERTILIZATION_JULIANDAY :-99. FERTILIZATION_HU :-99. !!endscheduledfertilization!! <endfertilizationdatabase> <endvegetationtype>