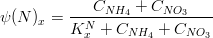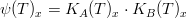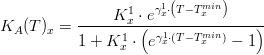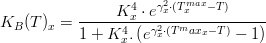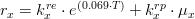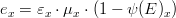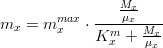Module MacroAlgae
From MohidWiki
Contents
Overview
Module MacroAlgae is a zero-dimensional biogeochemical model and was designed to compute the main biogeochemical processes related with macroalgae in aquatic environments. This module simulates two types of macroalgae:
- Attached macroalgae - macroalgae attached to the bottom of the water column
- Drifting macroalgae - macroalgae dettached from the bottom drifting in the water column
The evolution of the two macroalgae types is computed in the exactly same way (although different parameters are allowed for each type). The main differences relate to the fact that attached macroalgae are not transported by flow and drifting macroalgae are. On top of the main primary production limiting factors (e.g. light, nutrients, temperature), attached macroalgae growth is also limited by salinity (optionally), by suspended particulate matter deposition fluxes and also by shear stress near the bottom, which dettaches these macroalgae which become drifting macroalgae. Drifting macroalgae do not have this type of limitation (SPM deposition flux and obviously bottom shear stress) and when they reach the shore the only process computed is an increased mortality rate.
This module is independent of the pelagic and benthic biogeochemical model being used to compute other biogeochemical processes, thus it can be coupled with Module WaterQuality, Module CEQUALW2 or Module Life, or even Module Benthos.
One feature of this macroalgae model is that it is fully coupled with the 3D transport model, as macroalgae, although attached in the bottom, can influence water properties in the all water column and not only to the grid cell near the bottom (e.g. light attenuation, nutrients uptake, etc). This is the case when the macroalgae height is higher than the grid cell height. Basically, an occupation ratio is computed for each grid cell, based on the bottom distribution and macroalgae height.
Equations
As mentioned before, attached macroalgae evolution, in terms of biogeochemical processes, is computed exactly the same way as drifting macroalgae, and can be written as:
where,
*is the type of macroalgae (attached or drifting) *
is the macroalgae distribution *
is the gross production rate (in
) *
is the respiration rate (in
) *
is the excretions rate (in
) *
is the natural mortality rate (in
) *
is the lost chain grazing rate (in
)
Gross production rate
The gross production rate 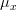 is given by:
is given by:
where,
*is the macroalgae maximum growth rate (in
) *
is the nitrogen limiting factor *
is the phosphorus limiting factor *
is the light limiting factor *
is the temperature limiting factor *
is the salinity limiting factor
Limiting factors
Nutrients
The nitrogen limiting factor can be computed as:
where,
*is the ammonia concentration in
*
is the nitrate concentration in
*
is the nitrogen half-saturation constant in
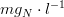
The phosphorus limiting factor can be computed as:
where,
*is the inorganic phosphorus concentration in
*
is the phosphorus half-saturation constant in
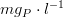
Light
Temperature
Macroalgae's temperature limiting factor is given by:
where,
Salinity
Respiration rate
The macroalgae respiration rate is given by:
where,
*is the macroalgae endogenous respiration (in
) *
is the water temperature (
) *
is the macroalgae photo respiration rate (in
)
Excretion rate
The macroalgae excretion rate is given by:
where,
*is the reference excretion rate (in
)
Natural mortality rate
The macroalgae natural mortality rate is given by:
where,
*is the maximum natural mortality rate (in
) *
is the half-saturation constant
Numerical scheme
Module MacroAlgae solves the differential equations described above using an Euler forward scheme (explicit method), which was used in order to make the code organization as simple as possible. When using this numerical method there are, nevertheless, some restraints in terms of temporal discretization, which can cause numerical instabilities that can be solved, at some extent, by using a smaller time step.
Only for a specific calculation (drifting macroalgae mortality when reaching the shore), an Euler backward scheme (implicit method) is used to insure numerical stability.
Module MacroAlgae has an independent time step in relation to the model’s main time step. Normally the time step to integrate the differential equations describing these kind of biogeochemical reactions is much higher than the time step used to solve hydrodynamic and transport equations, and as the set of equations in this module is quite simple, there is no significant increase in terms of computational costs when using a smaller time step.
User guide
Waterproperties data file options
In order to activate this module, specify in Module WaterProperties input data file, in each property involved in macroalgae processes the following keyword:
MACROALGAE : 1
Here is the property list which has to be with the macroalgae processes keyword activated:
macroalgae drifting macroalgae oxygen ammonia* nitrate* dissolved non-refractory organic nitrogen* particulate organic nitrogen* inorganic phosphorus** dissolved non-refractory organic phosphorus** particulate organic phosphorus**
*if computing the nitrogen cycle
**if computing the phosphorus cycle
The following step, still in Module WaterProperties input data file, in the header section, the following keywords:
MACROALGAE_MASS : 0.001 !macroalgae bottom distribution in gC/m2 MACROALGAE_HEIGHT : 0.25 !macroalgae reference height in meters
Ultimately one must parameterize the influence of macroalgae in light extincion. Thus, in the macroalgae property block the following keywords must be defined:
LIGHT_EXTINCTION : 1 !activates macroalgae influence on light extinction EXTINCTION_PARAMETER : 0.01 !defines the extinction parameter for macroalgae
Remember that in property macroalgae advection-diffusion should be off:
ADVECTION_DIFFUSION : 0
Property drifting macroalgae advection-diffusion should be on.
Macroalgae data file options
Then, activate Module MacroAlgae in MOHID GUI as described here, which enables to edit the MacroAlgae input data file. In this file, data is stored and organized in keywords and blocks. The main keywords are:
Keyword Default value Description DT : 3600] !Time step compute biogeochemical processes PELAGIC_MODEL : WaterQuality !Defines which pelagic biogeochemical model is coupled NITROGEN : 0 !Compute the nitrogen cycle PHOSPHORUS : 0 !Compute the phosphorus cycle
For each macroalgae type a block must be specified:
Attached macroalgae - <begin_macroalgae> ... <end_macroalgae> Drifting macroalgae - <begin_driftingmacroalgae> ... <end_driftingmacroalgae>
Inside each block specific parameters are defined:
Keyword Default value Description GROWMAX : 0.4 !maximum growth rate TOPTMIN : 20. !optimum minimum temperature for growth TOPTMAX : 25. !optimum maximum temperature for growth TMIN : 5 !minimum temperature for growth TMAX : 40. !maximum temperature for growth TCONST1 : 0.05 !constant to control temperature response curve shape TCONST2 : 0.98 !constant to control temperature response curve shape TCONST3 : 0.98 !constant to control temperature response curve shape TCONST4 : 0.02 !constant to control temperature response curve shape PHOTOIN : 90. !optimum radiation value ENDREPC : 0.009 !endogenous respiration rate PHOTORES : 0.018 !photorespiration rate EXCRCONS : 0.008 !excretion rate MORTMAX : 0.003 !natural mortality rate MORTCON : 0.03 !mortality half saturation constant GRAZCONS : 0.00008 !grazing rate over macroalgae SOLEXCR : 0.25 !fraction of soluble inorganic material excreted by macroalgae DISSDON : 0.25 !fraction of dissolved organic material excreted by macroalgae NSATCONS : 0.065 !nitrogen half-saturation constant for macroalgae PSATCONS : 0.001 !phosphorus half-saturation constant for macroalgae RATIONC : 0.18 !macroalgae nitrogen/carbon ratio RATIOPC : 0.024 !macroalgae phosphorus/carbon ratio MACROALGAE_MINCONC : 1e-12 !minimum residual value for macroalgae abundance MIN_OXYGEN : 1e-8 !minimum oxygen concentration for macroalgae growth DEPLIM : 5e-6 !maximum SPM deposition flux for macroalgae growth(kg m-2 s-1)* EROCRITSS : 0.1 !critical shear stress to occur macroalgae dettachment (in Pa)* SALT_EFFECT : 0 !include salinity limitation on macroalgae growth SALTOPT : 20 !macroalgae optimum salinity for growth SALTCRIT : 5. !macroalgae critical salinity limit growth SALTMIN : 0. !macroalgae minimum salinity for growth SALTMAX : 45. !macroalgae maximum salinity for growth BEACHED_MORT_RATE : 0.01 !beached drifting macroalgae mortality rate**
*only for attached macroalgae
**only for drifting macroalgae
Performing outputs of biogeochemical rates
In order to perform outputs of biogeochemical rates and other parameters of the MacroAlgae model, you can define in the WaterProperties input data file the following options.
BOXFLUXES : ..\..\GeneralData\Boxes.dat !path to a boxes data file
The same keyword, referencing the same boxes data file must be defined in the Hydrodynamic input data file. This insures the output of the water.BXM file, which contains the spatial integration of the water volumes into the previously boxes defined.
Also at least one water property needs to have the following keyword activated:
BOX_TIME_SERIE : 1
Next step is to define the rates which the user wants to analyse. This is done similarly to the other biogeochemical modules. All exchange rates are available for output (e.g. macroalgae to ammonia, ammonia to macroalgae, oxygen to macroalgae, macroalgae to oxygen, etc...)
This can be done by defining in WaterProperties input data file, a block as the one presented below:
<beginwqrate> NAME : macroalgae ammonia uptake DESCRIPTION : what goes from ammonia to macroalgae FIRSTPROP : ammonia SECONDPROP : macroalgae MODEL : MacroAlgae <endwqrate>
<beginwqrate> NAME : macroalgae ammonia excretions DESCRIPTION : what goes from macroalgae to ammonia FIRSTPROP : macroalgae SECONDPROP : ammonia MODEL : MacroAlgae <endwqrate>
One can also analyse other parameters such as:
- macroalgae gross production
- macroalgae temperature limiting factor
- macroalgae light limiting factor
- macroalgae nutrients limiting factor
- macroalgae nitrogen limiting factor
- macroalgae phosphorus limiting factor
- macroalgae salinity limiting factor
In these cases, in the <beginwqrate>...<endwqrate> block, the FIRSTPROP keyword relates to the parameter to be analysed and the SECONDPROP relates to the type of macroalgae. For example, if one wishes to analyse the gross production of attached macroalgae and drifting macroalgae, the data file should look like this:
<beginwqrate> NAME : macroalgae gross production DESCRIPTION : macroalgae gross production FIRSTPROP : grossprod SECONDPROP : macroalgae MODEL : MacroAlgae <endwqrate>
<beginwqrate> NAME : macroalgae gross production DESCRIPTION : macroalgae gross production FIRSTPROP : grossprod SECONDPROP : drifting macroalgae MODEL : MacroAlgae <endwqrate>
In the end a time series file with an *.BXM extension and with the name of the rate (set by keyword 'NAME'), is written.
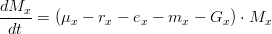
![\mu _x = \mu^{\max }_x \cdot \min \left[{\psi (N)_x ,\psi (P)_x} \right]\cdot \psi (E)_x \cdot \psi (T)_x \cdot \psi (S)_x](/images/math/3/0/b/30be434a3eddb53c2e52bd1412237614.png)